Videopod
The Quest for Next-Generation Covalent Drugs: Mapping Unexplored Molecular Targets
Audiopod
Beyond Conventional Drug Targets: Unraveling New Avenues for Covalent Drug Development
Synopsis
Breaking Boundaries: Discovering Unmapped Molecular Targets for Novel Covalent Drugs
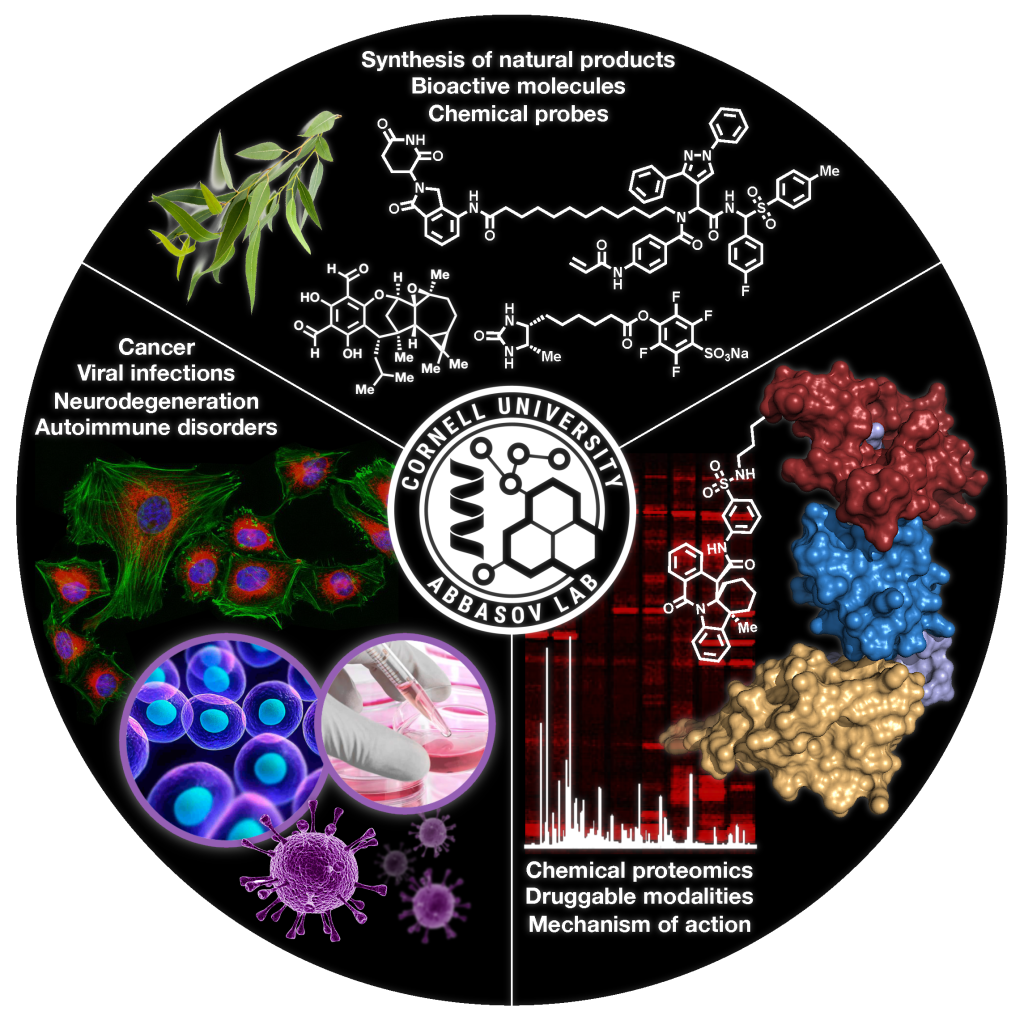
Covalent drugs are molecules that irreversibly bind to specific, targeted sites in the body. They work to inhibit the disease-causing functions of certain proteins by preventing them from interacting with other substances. This is a highly promising field of drug development and the focus of Dr Mikail Abbasov from Cornell University, New York, USA. By creating and utilising new technologies and through collaborative research, Dr Abbasov has mapped novel molecular targets for potential covalent drugs to treat ailments ranging from cancer to autoimmune diseases.
Covalent Drugs: Creating Irreversible Bonds
Therapeutic drugs come in numerous forms for an endless number of diseases. Depending on the ailment in question, a drug will target certain biochemical reactions in the body to reduce its negative impacts. This involves the drug in question binding to a target protein in cells. Whereas some therapeutics form bonds that are reversible, others form permanent bonds to create a long-lasting effect. For example, covalent drugs form strong, irreversible covalent bonds with proteins. Well-known examples of this type of drug include the anti-inflammatory, Aspirin, the antibiotic, Penicillin, and the cancer drug, Ibrutinib. In the past, there have been safety concerns surrounding covalent drugs, with many scientists viewing their irreversible binding actions as dangerous. Some studies showed that these drugs could be metabolised to create highly reactive intermediates that, if accidentally targeted the wrong protein, could become toxic to a patient. More recently, a resurgence in interest in covalent drugs has exploited their compelling benefits whilst fine-tuning how they react in the body to ensure their safety. These new drugs are called targeted covalent inhibitors and they work by first being guided towards and weakly bonding to the desired protein through a small pocket. This initial interaction facilitates the subsequent formation of the irreversible covalent bond with a nearby amino acid. Consequently, the function of that protein is inhibited. Covalent inhibitors target nucleophilic amino acids – molecules on proteins that form bonds with an electrophile by donating an electron pair. They can be designed to target very specific nucleophiles on a protein so that other proteins within the same family are not impacted when the drug is administered. This clever technique is being utilised in a number of drugs that are currently in late-stage development. Many of them are targeting different types of cancer and some are even being tested for COVID-19 treatments, such as Paxlovid. Although the field of covalent drugs is highly promising, there is a distinct lack of research as of yet and much more work is needed to make full use of its potential. Dr Mikail Abbasov from the Department of Chemistry and Chemical Biology at Cornell University in New York, USA dedicates his research to improving our knowledge and understanding of the targets of covalent drugs and how they could be used to create ground-breaking therapies. Working alongside his colleagues, Dr Abbasov explains, ‘Our highly interdisciplinary research program spans the fields of chemistry, biology and physics with an emphasis on the development of innovative chemical proteomic tools and technologies that modulate protein function and interrogate pathophysiological signalling pathways associated with human diseases, such as cancer, viral infections, autoimmune and neurodegenerative disorders. Findings from our laboratory have enriched our understanding of the mechanistic underpinnings of pathological processes and provide valuable leads for the design and development of novel therapeutics.'
Discovering Simple Molecules with Drug Potential
Historically, to understand the relationships between the structure and activity of a protein, total synthesis followed by structural derivatisation has been performed. Total synthesis is the process of using chemistry to form a complete, complex molecule using commercially-available ingredients. These complex molecules are usually natural products, which are substances that are created by living organisms and plants, meaning they are already found in nature. Structural derivatisation refers to chemically transforming a chemical compound into a different, yet structurally similar one, known as a derivative. However, Dr Abbasov saw that these synthetic techniques to create natural products were not usually guided by hypotheses that took into consideration the structural features of the compounds that are needed for bioactivity. Therefore, Dr Abbasov, in collaboration with Dr Daniel Romo at Baylor University in Texas, developed a new strategy for natural product synthesis which he called ‘pharmacophore-directed retrosynthesis’. A pharmacophore is the section of a molecule that is necessary for its biological and pharmacological activity. Retrosynthesis is a method used to understand molecule synthesis by breaking down the final product into more simple precursors. They are continually simplified until the resulting molecules are commercially available or easy to use in a synthetic reaction for the target molecule. Dr Abbasov performed pharmacophore-directed retrosynthesis with a pre-determined or a hypothetical pharmacophore of a natural product in mind. As a result, the intermediates that were synthesised in the process could each be tested for their bioactivity during the total synthesis of the natural product. Putting this innovative new method into practice, Dr Abbasov investigated gracilin A, a natural product isolated from the Mediterranean sponge Spongionella gracilis. The biological role of the gracilin A prior to this study was largely unexplored. For this reason, Dr Abbasov endeavoured to use his pharmacophore-directed retrosynthetic approach to create structurally simplified derivatives of gracillin A that can be accessed in the early stages of the synthesis. He aimed to identify derivatives with selective affinity (attraction) towards cyclophilin A and cyclophilin D, proteins associated with immunosuppressive and neuroprotective activities, respectively. Consequently, Dr Abbasov identified simplified, ultrapotent derivatives that exhibited activity greater than that of gracilin A. This is an important finding because it demonstrates that structurally simplified intermediates can possess greater biological activity than that of the parent natural product, thus reinforcing the advantages that this approach offers for the rapid discovery of potential therapeutics. In particular, the derivatives that have a high affinity for cyclophilin D over cyclophilin A could be useful as neuroprotective drugs that do not have immunosuppressive side effects.
Discovering and Mapping Druggable Lysines
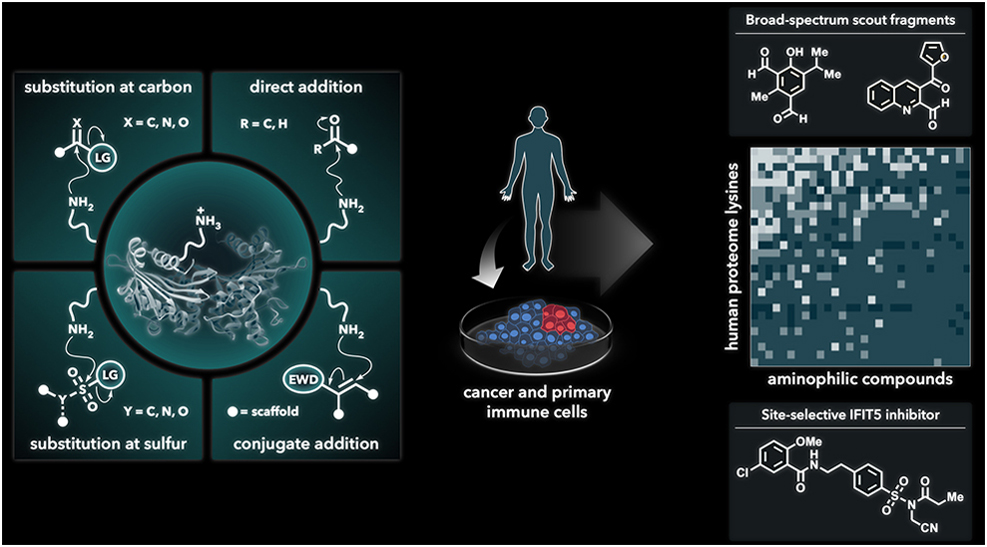
In a subsequent study, Dr Abbasov and his colleagues conducted new and exciting research on biological targets for covalent drugs. In particular, they focused on covalent targeting of amino acids – the building blocks of proteins. Twenty different amino acids are found in the human body and historically, two of them called cysteine and serine have been the main focus for covalent drugs. However, this limits the possibilities for drug discovery. Consequently, Dr Abbasov, in collaboration with Dr Benjamin Cravatt at Scripps Research in California, set out to investigate another amino acid called lysine. They also brought in teams from the Massachusetts Institute of Technology, Pfizer and Leiden University for the project. Together, they developed a chemoproteomic technology that allowed them to study how lysines react with small molecules. They aimed to find aminophilic electrophiles – chemical species that form irreversible bonds with nucleophilic amine groups on lysines. This ground-breaking work led Dr Abbasov to profile and log over 200 of these small molecules. By testing human cancer and immune cells, the team found thousands of reactive lysines. This included functional sites on proteins that were previously seen as too difficult to target. Collating this new data allowed Dr Abbasov and his colleagues to create a comprehensive map of lysines that are now known to be possible drug targets. By fine-tuning both the recognition portion and the reactive fragment of the small molecules, they were able to identify a wide range of proteins in which these lysines are found. These proteins have a wide range of functions in the human body and are known to be involved in various diseases, including, for example, enzymes and transcription factors (needed for protein synthesis). By inhibiting the function of disease-relevant proteins, these types of small molecules could serve as drugs to prevent the harmful progression of a variety of diseases. According to Dr Abbasov, ‘The findings from this project provide a blueprint for chemical biologists and medicinal chemists to understand the rules that govern lysine-targeting covalent inhibitor design, eventually leading to new treatments and drugs.’ To aid this development, after mapping the druggable lysines, Dr Abbasov and his team combined this data with human genetic information and cell activation measurements. This helped to refine their understanding of how different small molecules can impact the activity of the protein and consequently, the cell. They were able to demonstrate that, through lysine binding, aminophilic compounds disrupted a number of biochemical functions within cells, including those involved in the innate immune response. With such real promise for finding and creating new covalent drugs, Dr Abbasov’s work is not stopping there. ‘Findings from our laboratory have enriched our understanding of the mechanistic underpinnings of pathological processes and provide valuable leads for the design and development of novel therapeutics’ notes Dr Abbasov. And this research is ongoing; only one year after starting his position at Cornell University, Dr Abbasov secured a $2M grant from the National Institute of General Medical Sciences to continue discovering the druggable targets of the human body. Our highly interdisciplinary research program spans the fields of chemistry, biology and physics with an emphasis on development of innovative chemical proteomic tools and technologies that modulate protein function and interrogate pathophysiological signaling pathways associated with human diseases, such as cancer, autoimmune and neurodegenerative disorders. Our research program centers on the use of chemical biology approaches, including mass spectrometry-based chemoproteomics, and draws upon various disciplines such as cell and molecular biology, medicinal and organic chemistry, biochemistry, biophysics, and bioinformatics. Findings from our laboratory may enrich our understanding of the mechanistic underpinnings of pathological processes and provide valuable leads for the design and development of novel therapeutics.
Training
TRAINING
Students involved in our research program will engage in rigorous, highly interdisciplinary, and collaborative projects and will gain knowledge in: (1) organic chemistry with emphasis on total synthesis, chemical methodology and medicinal chemistry, (2) cutting-edge mass spectrometry-based chemoproteomic technologies with a special focus on untargeted and multiplexed methods, and (3) cellular and biochemical characterization of disease-relevant proteins and associated metabolic pathways in human cancer and immune cells. A knowledge base of organic synthesis and chemical proteomics acquired in our laboratory will enable students to approach biological problems from a unique perspective, with particular attention towards how molecular structure and reactivity affect biological systems. During the course of research, students in our laboratory develop the following skill set and expertise:
- Chemical synthesis: multistep synthesis with analytical characterization, natural product total synthesis, chemical methodology, probe development, structural optimization, structure-activity relationship (SAR) analysis.
- Cell culture: mammalian cell lines, bacterial and primary cultures.
- Proteomics: activity-based protein profiling (ABPP) technologies, including tandem mass tag (TMT), isotopic tandem orthogonal proteolysis (isoTOP), stable isotope labeling by amino acids in cell culture (SILAC), reductive dimethylation (ReDiMe).
- Molecular imaging: gel-based ABPP, confocal microscopy and flow cytometry.
- Biochemistry: protein preparation, overexpression, purification, and protein engineering.
- Molecular biology: cloning, transfections, PCR, RNAi, and CRISPR.

Projects
A GLIMPSE INTO OUR RESEARCH PROJECTS
(for a complete list of projects please contact Mikail)
- Small molecule proteases for targeted, proteasome-independent degradation of proteins in human cells
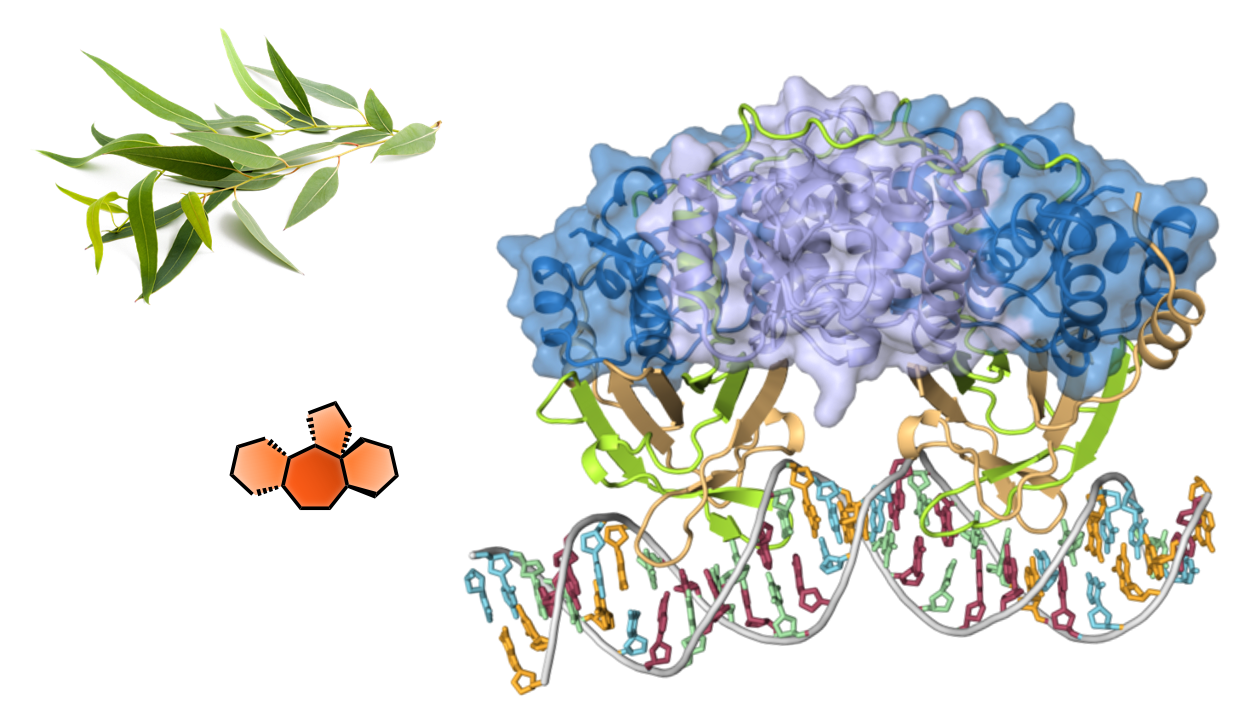
- Illuminating the druggable ribonucleoproteome with lysine-reactive meroterpenoid natural products
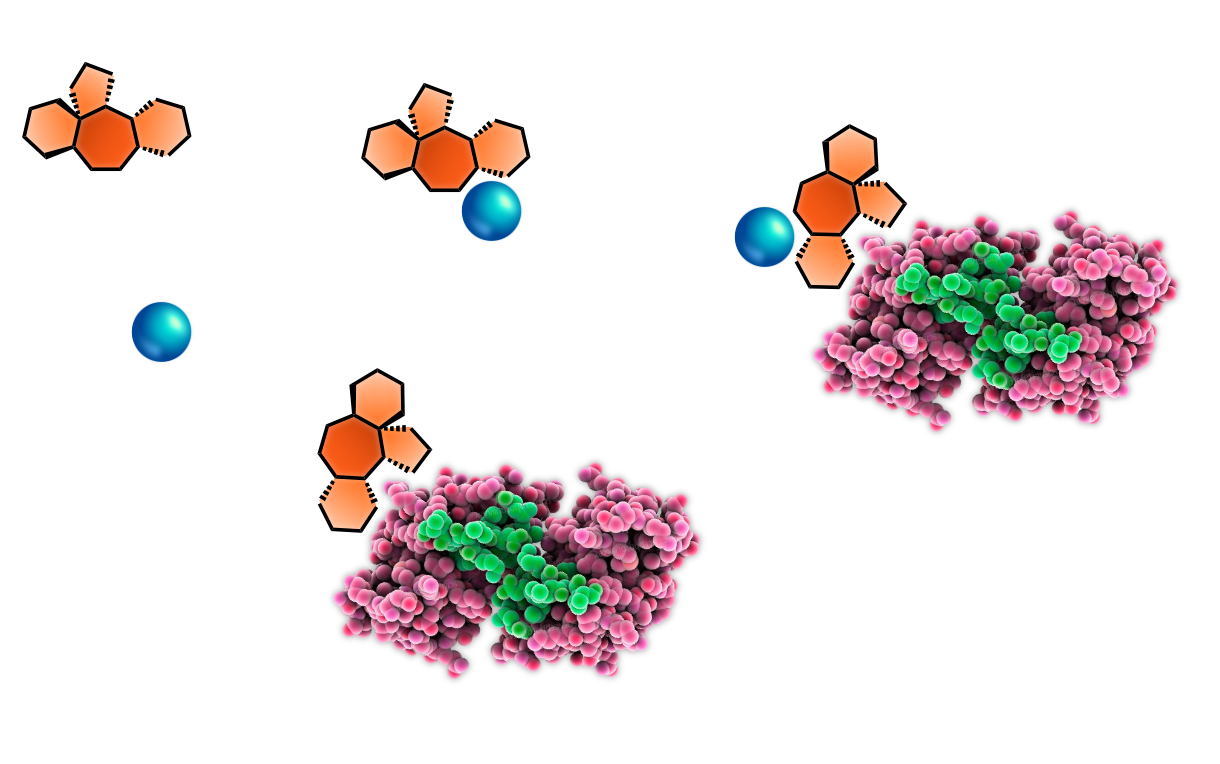
- Metal-responsive electrophiles for elucidating metal ion dyshomeostasis in native biological systems
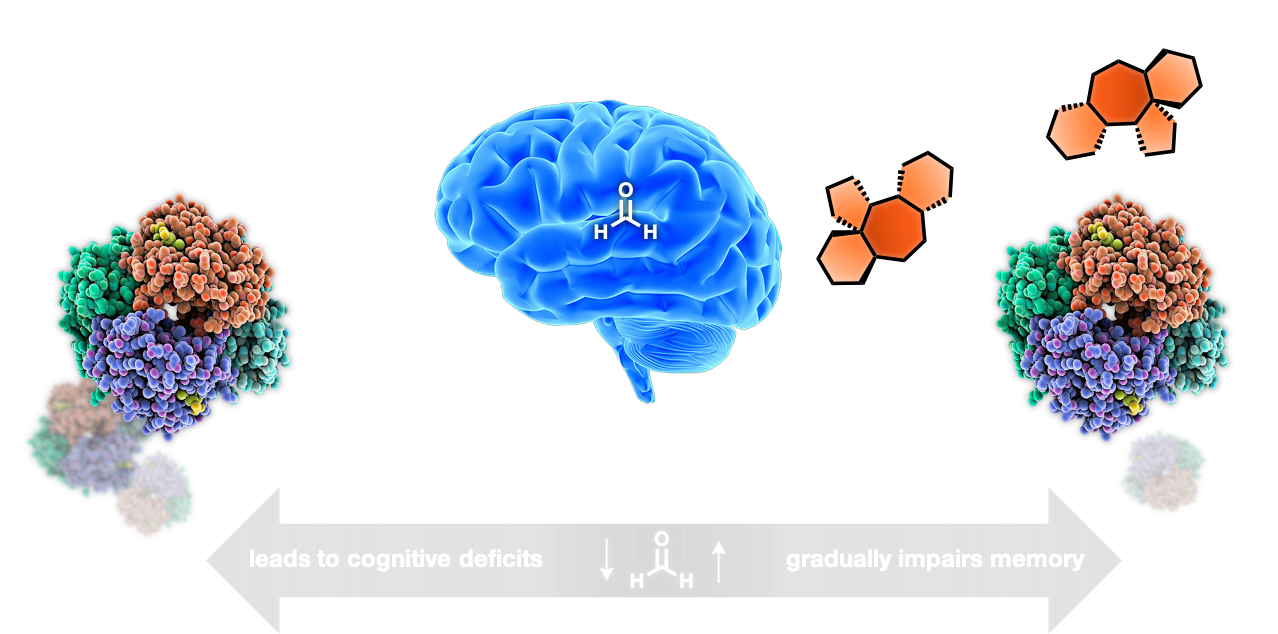
- Deciphering pathogenic dysregulation of endogenous formaldehyde with multicomponent reactions
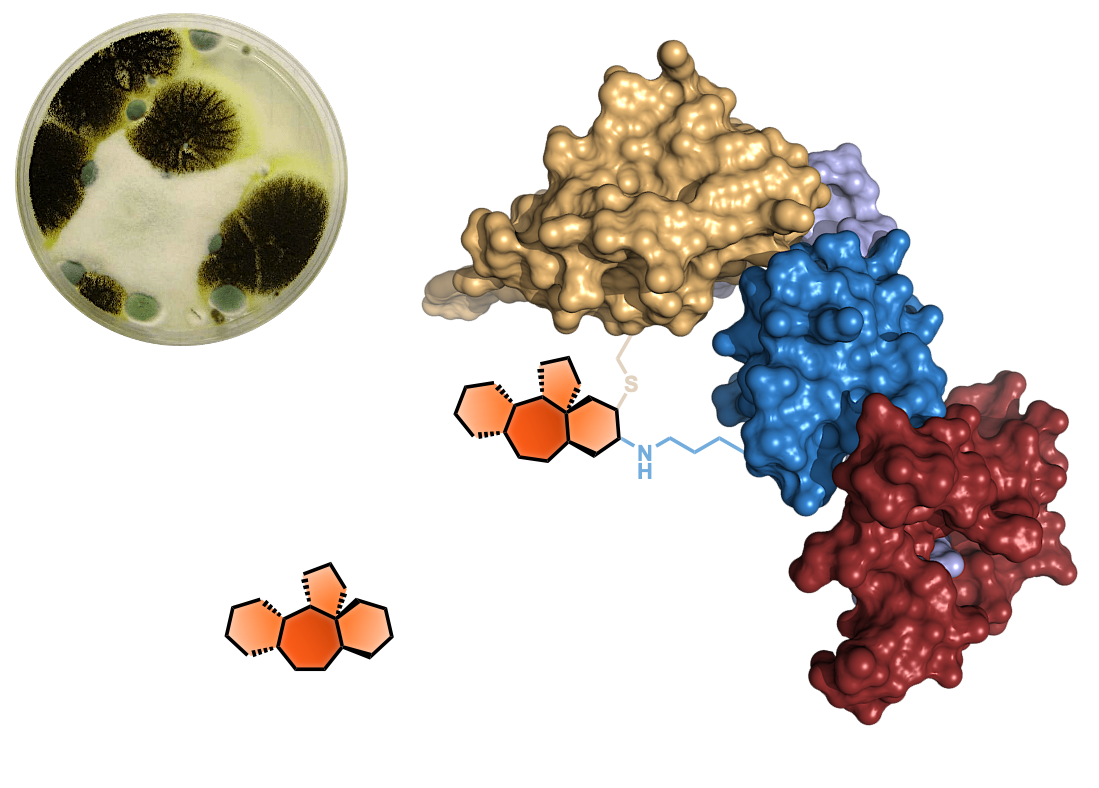
- Stachybotrys microspora secondary metabolites as chemoselective biselectrophiles for mapping proteinaceous cysteine-lysine clusters

Funding
2021–2024, Cornell University, Startup Funds

2021–2026, NIH/NIGMS MIRA (R35)

2022–2023, Adam and Rachel Broder Fund for Cancer Research
